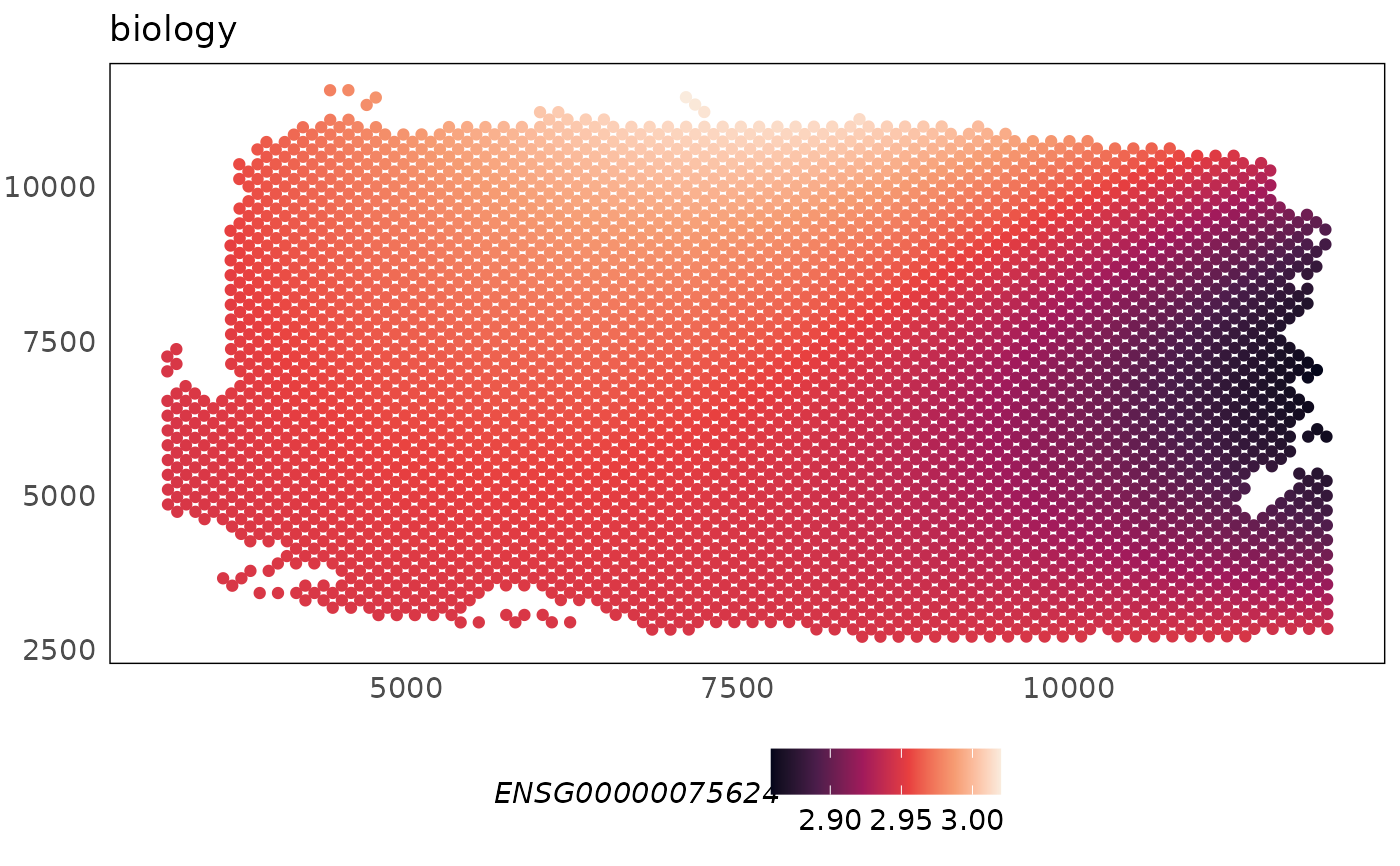
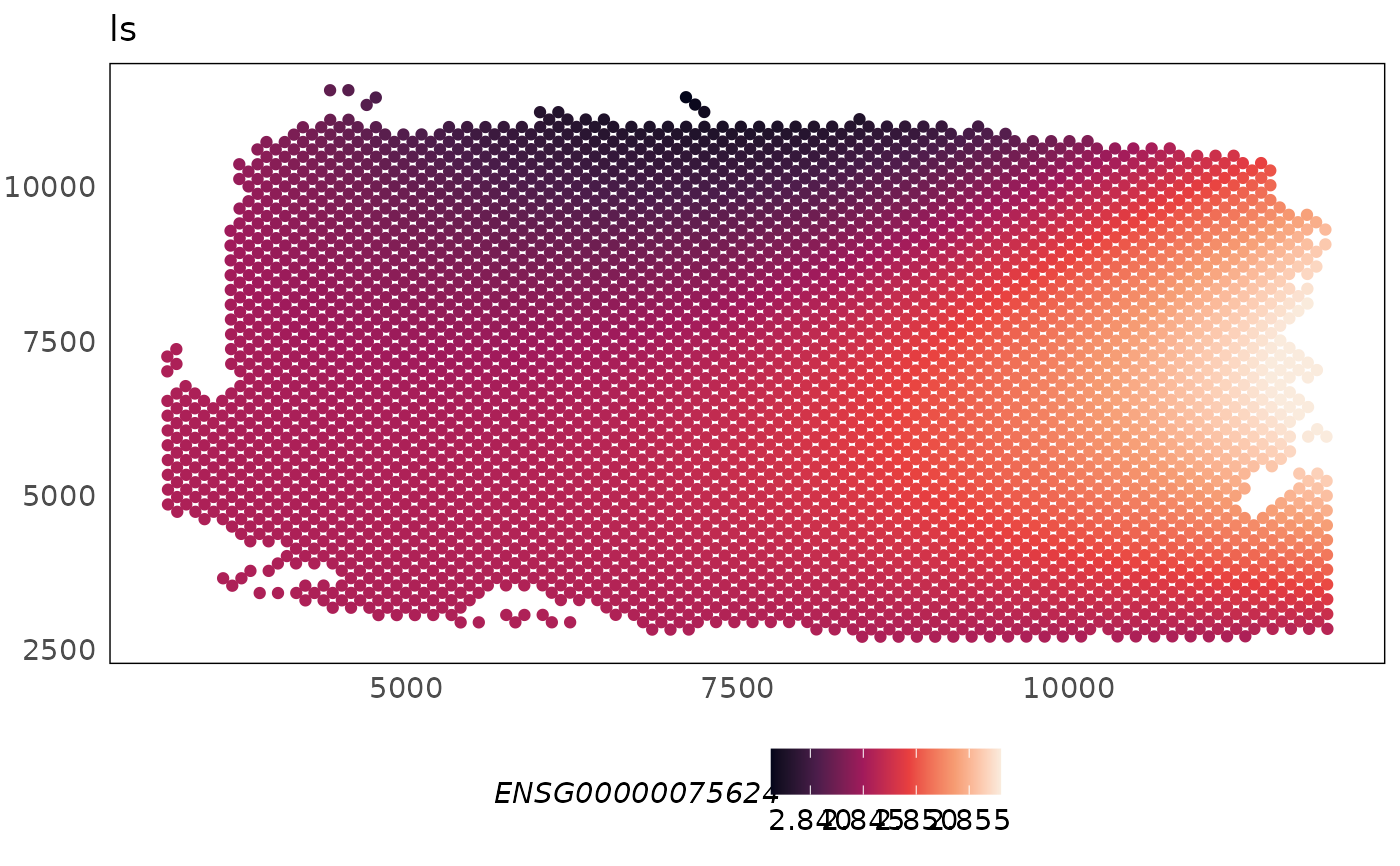
This function can be used to spatially visualise the library size, biology or batch specific effect modelled for each gene.
Usage
plotCovariate(spe, covariate = c("biology", "ls", "batch"), ...)Arguments
- spe
a SpatialExperiment object.
- covariate
a character, specifying the type of covariate to be plot: "biology" (default), "ls" to plot the library size effect, and "batch" to plot the batch-specific effect.
- ...
additional parameters to be passed to the plotSpatial function.
Examples
library(SpatialExperiment)
library(ggplot2)
data(HumanDLPFC)
# \donttest{
HumanDLPFC = SpaNorm(HumanDLPFC, sample.p = 0.05, df.tps = 2, tol = 1e-2)
#> (1/2) Fitting SpaNorm model
#> 201 cells/spots sampled to fit model
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 1, estimating gene-wise dispersion
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 1, log-likelihood: -1175583.616527
#> iter: 1, fitting NB model
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 1, iter: 1, log-likelihood: -1175583.616527
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 1, iter: 2, log-likelihood: -834517.676172
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 1, iter: 3, log-likelihood: -743559.059971
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 1, iter: 4, log-likelihood: -727273.138763
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 1, iter: 5, log-likelihood: -724794.857624
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 1, iter: 6, log-likelihood: -724387.227124
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 1, iter: 7, log-likelihood: -724302.329293
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 1, iter: 8, log-likelihood: -724278.669652 (converged)
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 2, estimating gene-wise dispersion
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 2, log-likelihood: -723927.814979
#> iter: 2, fitting NB model
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 2, iter: 1, log-likelihood: -723927.814979
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 2, iter: 2, log-likelihood: -723792.325311
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 2, iter: 3, log-likelihood: -723784.166135 (converged)
#> Hint: To use tensorflow with `py_require()`, call `py_require("tensorflow")` at the start of the R session
#> iter: 3, log-likelihood: -723784.166135 (converged)
#> (2/2) Normalising data
# plot spatial region annotations
p1 <- plotCovariate(HumanDLPFC, covariate = "biology", colour = ENSG00000075624) +
scale_colour_viridis_c(option = "F")
p1
 p2 <- plotCovariate(HumanDLPFC, covariate = "ls", colour = ENSG00000075624) +
scale_colour_viridis_c(option = "F")
p2
p2 <- plotCovariate(HumanDLPFC, covariate = "ls", colour = ENSG00000075624) +
scale_colour_viridis_c(option = "F")
p2
 # }
# }