Plot spatial transcriptomic annotations per spot
Usage
plotSpatial(
spe,
what = c("annotation", "expression", "reduceddim"),
...,
assay = SummarizedExperiment::assayNames(spe),
dimred = SingleCellExperiment::reducedDimNames(spe),
img = FALSE,
crop = FALSE,
imgAlpha = 1,
rl = 1,
circles = FALSE
)Arguments
- spe
a SpatialExperiment object.
- what
a character, specifying what aspect should be plot, "annotation", "expression", or reduced dimension ("reduceddim").
- ...
additional aesthetic mappings or fixed parameters (e.g., shape = ".").
- assay
a character or numeric, specifying the assay to plot (default is the first assay).
- dimred
a character or numeric, specifying the reduced dimension to plot (default is the first reduced dimension).
- img
a logical, indicating whether the tissue image (if present) should be plot (default = FALSE).
- crop
a logical, indicating whether the image should be cropped to the spatial coordinates (default = FALSE).
- imgAlpha
a numeric, specifying the alpha value for the image (default = 1).
- rl
a numeric, specifying the relative size of the text (default = 1).
- circles
a logical, indicating whether the spots should be plotted as circles (default = FALSE). This can be slower for large datasets.
Examples
library(SpatialExperiment)
library(ggplot2)
# load data
data(HumanDLPFC)
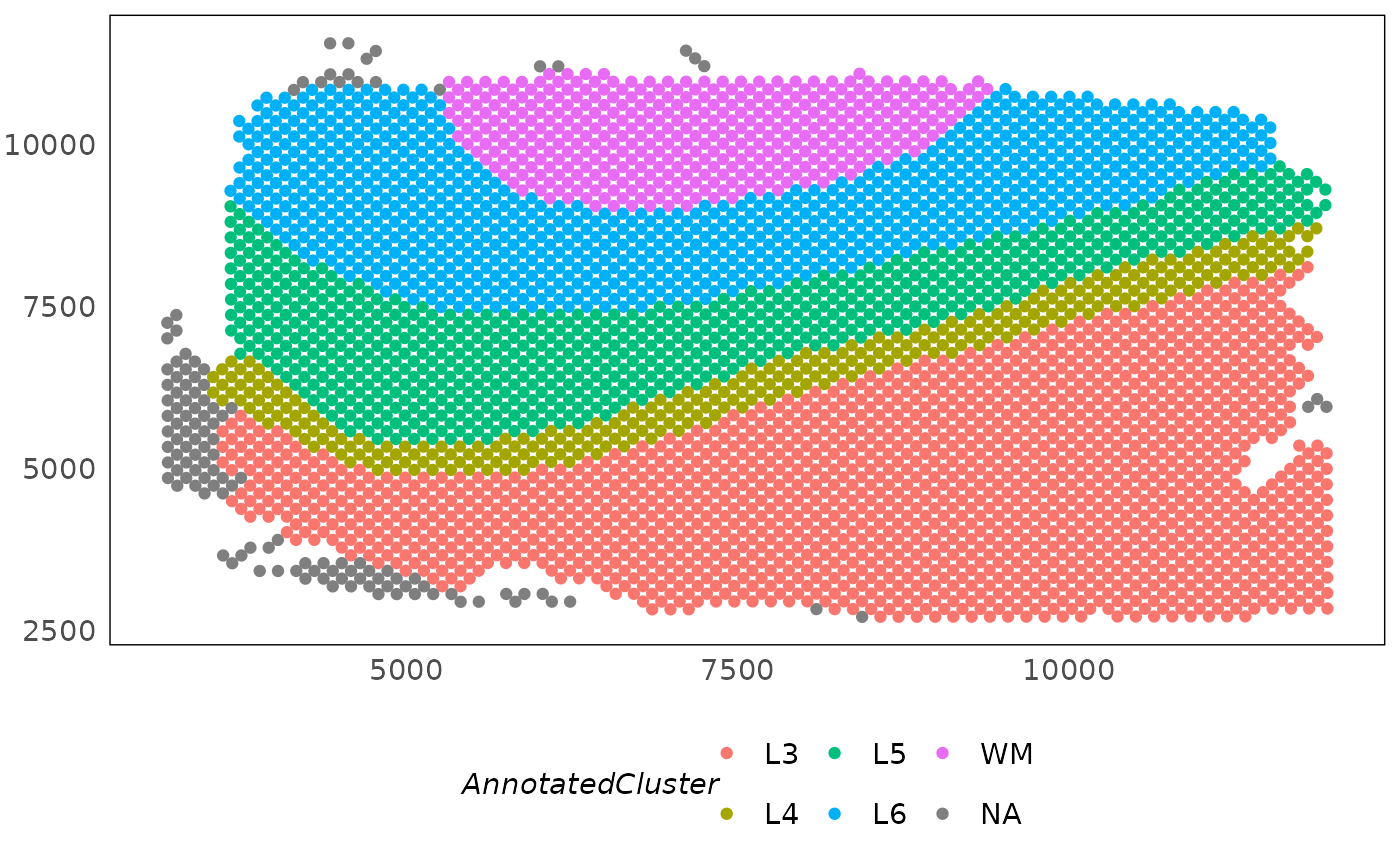
# plot spatial region annotations
p1 <- plotSpatial(HumanDLPFC, colour = AnnotatedCluster)
p1
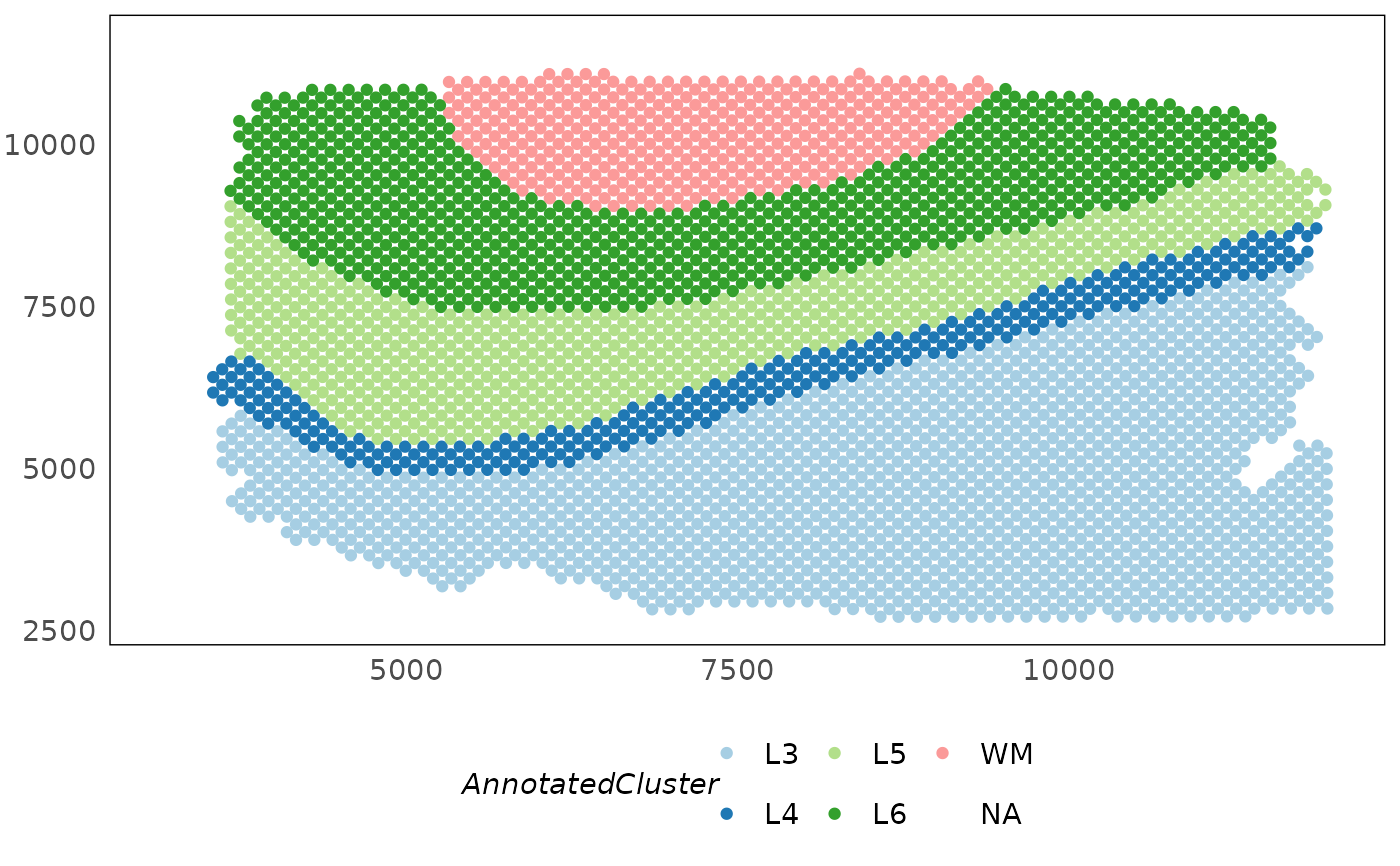
 # change colour scale
p1 + scale_colour_brewer(palette = "Paired")
#> Warning: Removed 127 rows containing missing values or values outside the scale range
#> (`geom_point()`).
# change colour scale
p1 + scale_colour_brewer(palette = "Paired")
#> Warning: Removed 127 rows containing missing values or values outside the scale range
#> (`geom_point()`).
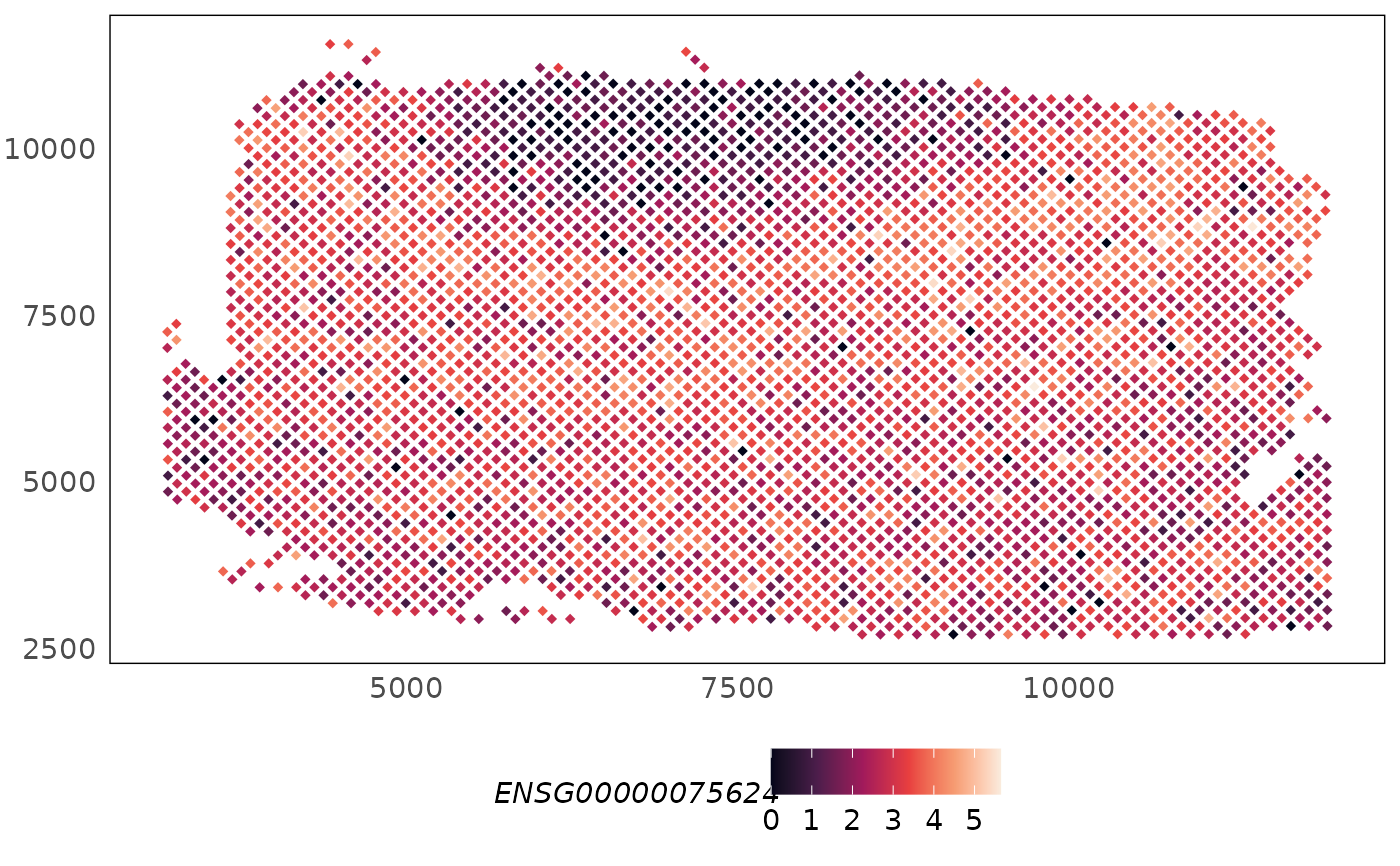
 # plot spatial expression
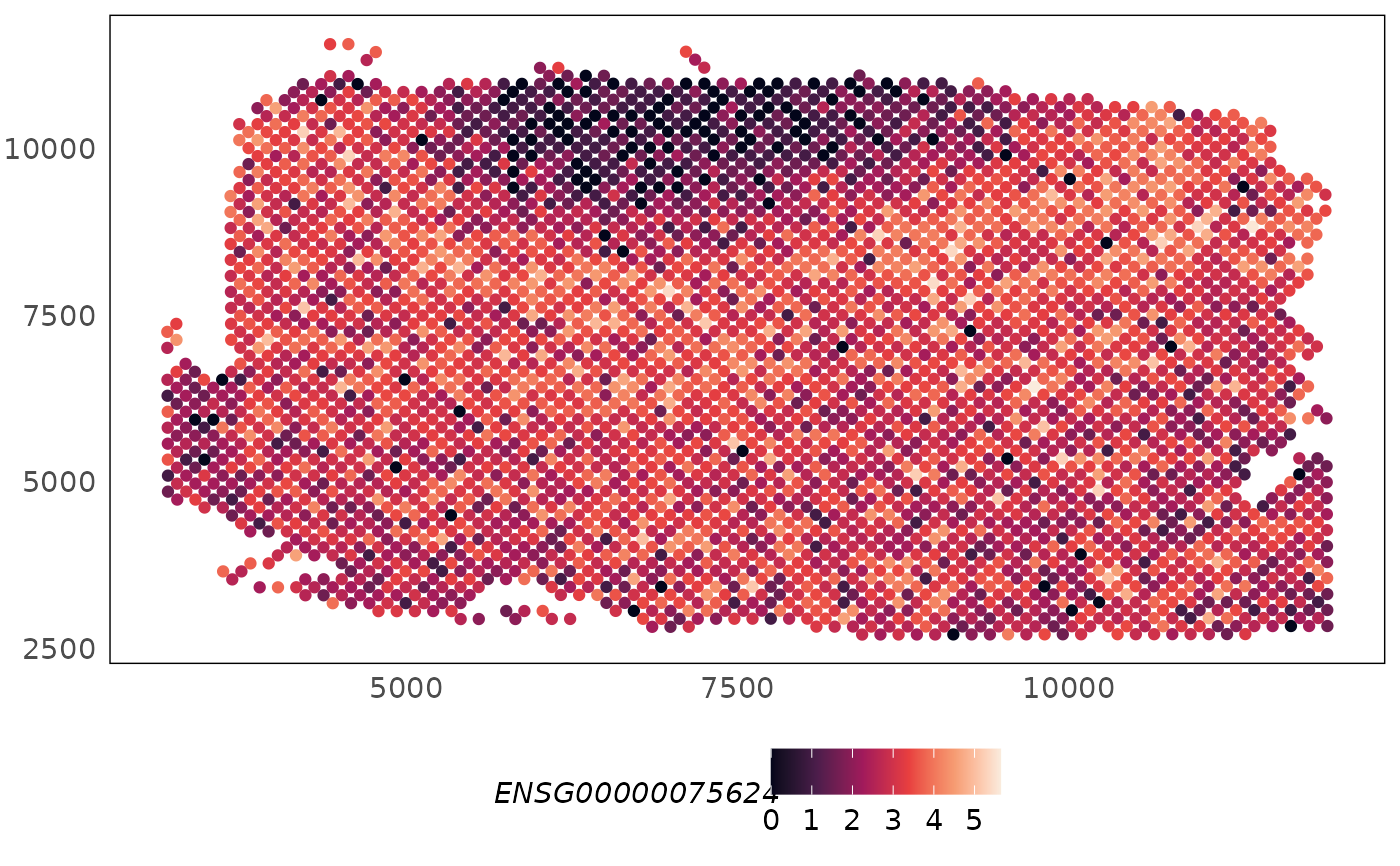
plotSpatial(HumanDLPFC, what = "expression", colour = ENSG00000075624) +
scale_colour_viridis_c(option = "F")
# plot spatial expression
plotSpatial(HumanDLPFC, what = "expression", colour = ENSG00000075624) +
scale_colour_viridis_c(option = "F")
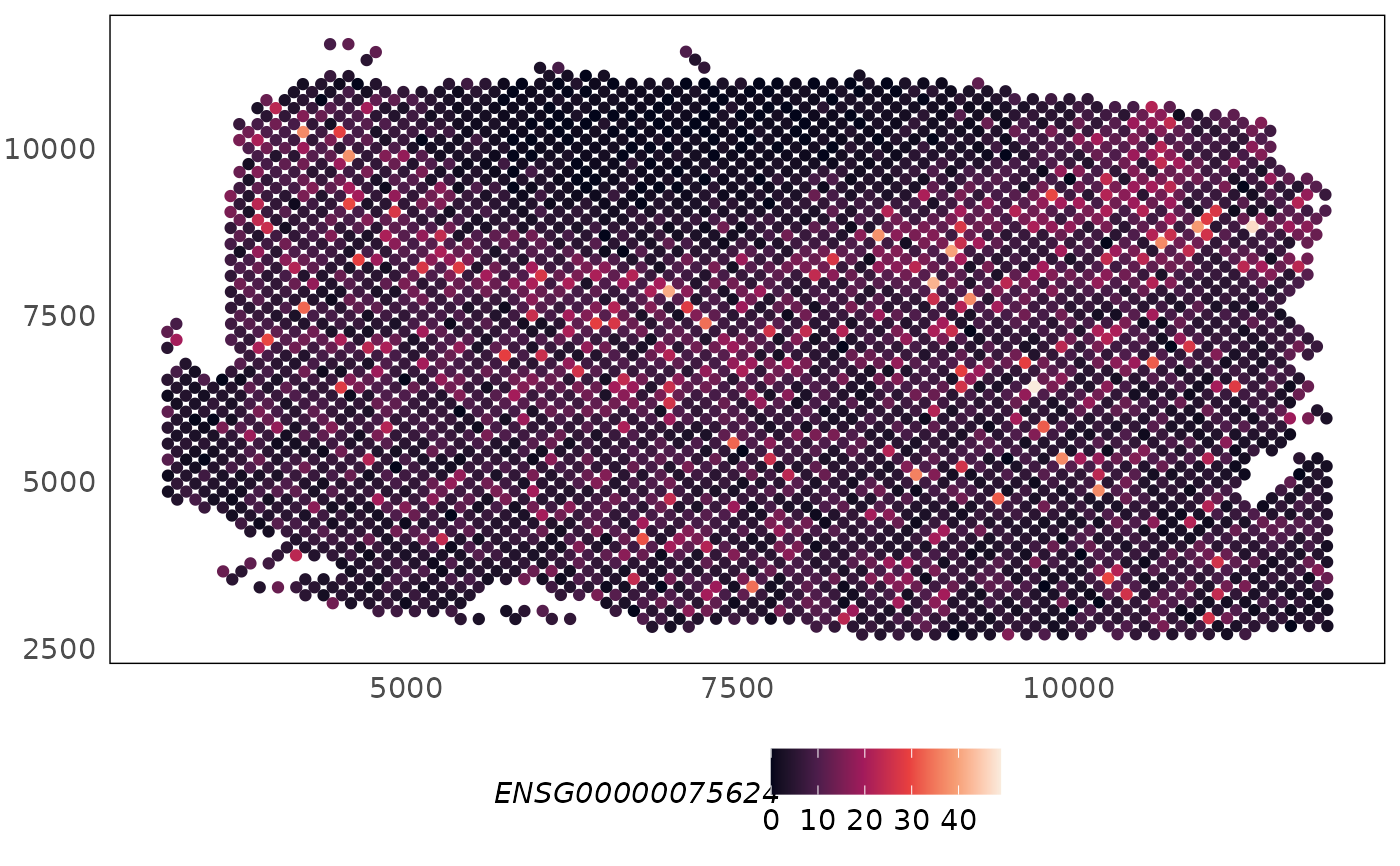 # plot logcounts
logcounts(HumanDLPFC) <- log2(counts(HumanDLPFC) + 1)
plotSpatial(HumanDLPFC, what = "expression", colour = ENSG00000075624, assay = "logcounts") +
scale_colour_viridis_c(option = "F")
# plot logcounts
logcounts(HumanDLPFC) <- log2(counts(HumanDLPFC) + 1)
plotSpatial(HumanDLPFC, what = "expression", colour = ENSG00000075624, assay = "logcounts") +
scale_colour_viridis_c(option = "F")
 # change point shape
plotSpatial(HumanDLPFC, what = "expression", colour = ENSG00000075624, assay = "logcounts", shape = 18) +
scale_colour_viridis_c(option = "F")
# change point shape
plotSpatial(HumanDLPFC, what = "expression", colour = ENSG00000075624, assay = "logcounts", shape = 18) +
scale_colour_viridis_c(option = "F")