Biography
Dharmesh D Bhuva is an early career computational biology researcher (PhD in 2020) in the Davis laboratory at SAiGENCI. He has developed high quality bioinformatics software and methods that enable exploration of molecular phenotypes in cancer systems. He currently works on developing statistical and computational methods to analyse high throughput spatial transcriptomics datasets.
Download my resumé .
- Spatial Statistics
- Cancer Systems Biology
- Computational Biology
PhD in Mathematics and Statistics, 2020
The University of Melbourne
MSc in Bioinformatics, 2015
The University of Melbourne
BSc in Computer Science, 2013
The University of Southampton
Experience
Responsibilities include:
- Development of novel spatial statistics methods to analyse sub-cellular spatial ‘omics data.
- Development of machine learning pipelines to automate analysis of histopathology images.
- Student supervision
Responsibilities include:
- Analysis of spatial and single-cell transcriptomics to identify mechanisms of drug-induced phenotypic plasticity in cancer.
- Data analysis for commercial research collaboration between the CRC for Cancer Therapeutics and Pfizer Inc.
- Analysis of generic and drug specific regulatory mechanisms in biological systems using data integration approaches.
- Analysis of multi-omics data associated with drug response across a diverse set of biological models including cell lines and patients.
Projects
Recent & Upcoming Talks
Featured Publications
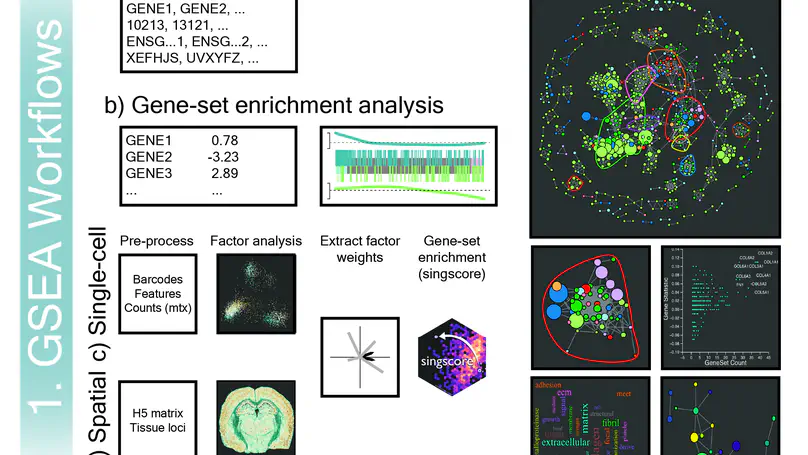
vissE.Cloud is a web-server that can help identify, visualise and explore higher-order biological processes from the results of a gene-set enrichment analysis (GSEA). It provides end-to-end analysis of scRNA-seq (10x Chromium) and spatial transcriptomics (10x Visium, 10x Xenium, and NanoString CosMx) datasets.
Recent Publications
Recent softwares
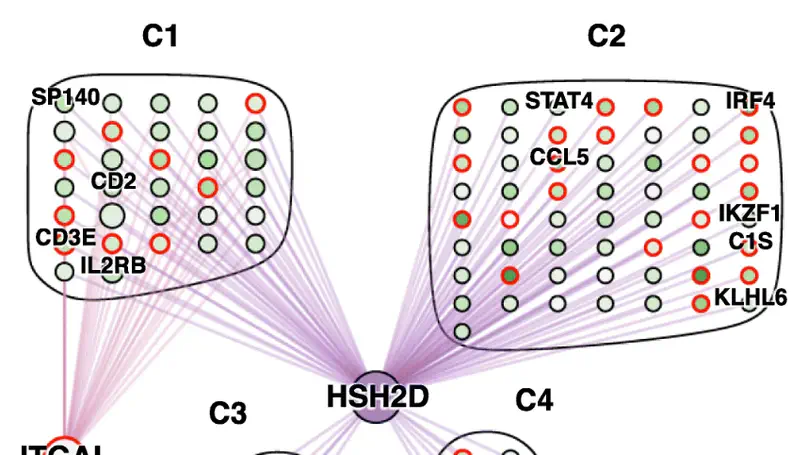
This package implements methods and an evaluation framework to infer differential co-expression/association networks. Various methods are implemented and can be evaluated using simulated datasets. Inference of differential co-expression networks can allow identification of networks that are altered between two conditions (e.g., health and disease).

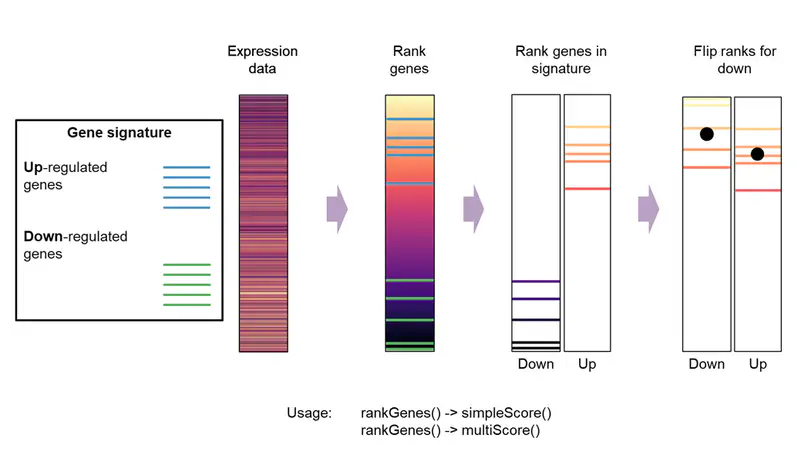
A simple single-sample gene signature scoring method that uses rank-based statistics to analyze the sample’s gene expression profile. It scores the expression activities of gene sets at a single-sample level.
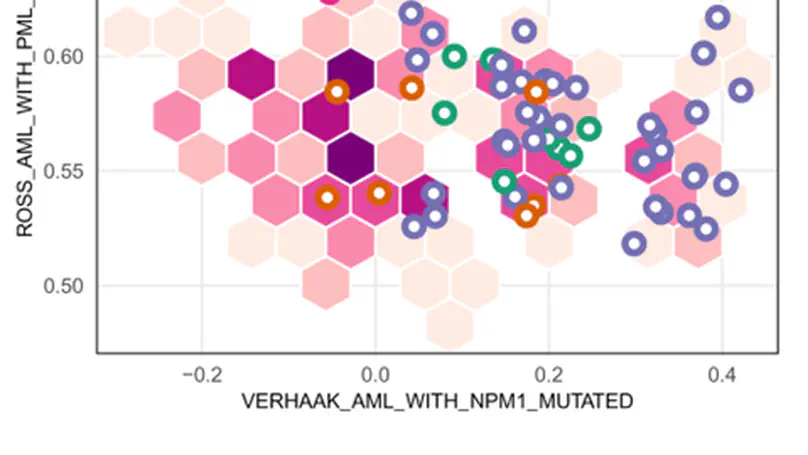
This workflow package shows how transcriptomic signatures can be used to infer phenotypes. The workflow begins by showing how the TCGA AML transcriptomic data can be downloaded and processed using the TCGAbiolinks packages. It then shows how samples can be scored using the singscore package and signatures from the MSigDB. Finally, the predictive capacity of scores in the context of predicting a specific mutation in AML is shown.The workflow exhibits the interplay of Bioconductor packages to achieve a gene-set level analysis.
standR is an user-friendly R package providing functions to assist conducting good-practice analysis of Nanostring’s GeoMX DSP data. All functions in the package are built based on the SpatialExperiment object, allowing integration into various spatial transcriptomics-related packages from Bioconductor. standR allows data inspection, quality control, normalization, batch correction and evaluation with informative visualizations.

This package enables the interpretation and analysis of results from a gene set enrichment analysis using network-based and text-mining approaches. Most enrichment analyses result in large lists of significant gene sets that are difficult to interpret. Tools in this package help build a similarity-based network of significant gene sets from a gene set enrichment analysis that can then be investigated for their biological function using text-mining approaches.
Contact
- test@example.org
- 888 888 88 88
- 450 Serra Mall, Stanford, CA 94305
- Enter Building 1 and take the stairs to Office 200 on Floor 2
-
Monday 10:00 to 13:00
Wednesday 09:00 to 10:00 - Book an appointment
- DM Me
- Zoom Me